For up to 500 injections - Retention time normalization kit for time-resolved mass spectrometry.
The iRT Kit allows standardized quality control of LC-MS platforms in up to 500 injections.
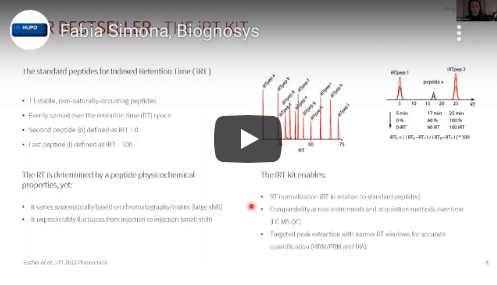
Indexed Retention Time (iRT) is a dimensionless value that defines the chromatographic retention of a peptide for a defined resin type (e.g. C18) relative to the iRT Standard. This concept that you can benefit from with the iRT Kit allows the development of standardized QC methods across different LC-MS instrument types.
The iRT Kit contains eleven non-naturally occurring synthetic peptides in a ready-to-use pooled mix. Peptides have been carefully optimized for stability, sensitivity, and even retention time spacing over the productive gradient. Moreover, the iRT Kit peptides spread in the ion mobility dimension which allows monitoring shifts and ion mobility space for timely calibration.
Balanced intensities of peptide signals minimize the possibility of undetected peptides under suboptimal LC conditions and make it easier to monitor any part of the gradient.
The iRT Kit peptides are AAA quantified, providing accurate information of peptide amounts and fully enabling their use for quantitative quality control.
In contrast to empirical retention time, the iRT value of a peptide is stable and enables accurate prediction of peptide retention for any chromatographic setup.
The iRT Kit allows straightforward determination of peptide iRT values and calibration of chromatographic systems.
By calibrating your LC system with the iRT Kit you will dramatically increase the throughput of your targeted proteomics (MRM and PRM) experiments – even up to 20 times – by bundling more transitions into one run.
The iRT peptides work with all Biognosys software solutions: Spectronaut® for DIA data analysis, SpectroDive™ for targeted proteomics experiments, SpectroMine™ for isobaric labeled quantitative experiments, and QuiC™ for MS performance monitoring.
The iRT Kit contains:
Dissolution Buffer (1x 2.0 ml tube)
iRT Standard (1x 2.0 ml tube)
The kit components are sufficient for approximately 500 injections depending on sample volume and injection amount.
The 2xiRT Kit contains:
Dissolution Buffer (2x 2.0 ml tube)
iRT Standard (2x 2.0 ml tube)
The kit components are sufficient for approximately 1000 injections depending on sample volume and injection amount.
The iRT Kit is used in time resolved mass spectrometry as a two-step process:
Determine iRT values of target peptides in relation to the iRT Standard
Calibrate LC system with iRT Standard and accurately predict retention time of target peptides