Proteomics is relevant not only to human health and disease, but also to animal species. In an exciting collaboration with the Leibniz Institute for Zoo and Wildlife Research (IZW) in Berlin, Germany, and the National Institute of Standards and Technology (NIST) in Charleston, USA, we profiled the blood proteome of 45 wild cheetahs from Namibia, Africa, to gain new insights into their underlying molecular biology and understand how male and female cheetahs differ.
Author: Roland Bruderer
Mass spectrometry proteomics is fast becoming a popular tool in clinical research, where it can be used across all stages of research from target discovery to clinical trials. However, it also has huge potential for fundamental biology in a wide range of species.
While most often applied to laboratory animals, proteomics can also be used for animal research in general, for which much less is known compared with well-characterized lab species. In fact, mass spectrometry proteomics is ideally suited for studies of animals, where the target proteins may be entirely uncharacterized.
Unlike affinity-based proteomics approaches, which rely on specific antibodies or aptamers, mass spectrometry is all inclusive and doesn’t require prior knowledge or reagents for the proteins of interest. It can also go deep into the entire proteome and is quantitative and reproducible at scale from tens to thousands of samples, allowing researchers to analyze and compare samples from multiple specimens and time points.
We recently demonstrated the power of Biognosys’ mass spectrometry proteomics technology for animal research in an exciting new study looking at the proteome of wild cheetahs living in Namibia in Southern Africa.
Not all cheetahs are alike
Although the behavior and ecology of cheetahs have been well studied, we know comparatively little about their underlying molecular biology. This is particularly true for their proteome composition, which has been virtually unexplored until now.
To understand more about the molecular biology of wild cheetahs, we brought our proteomics expertise to the Departments of Evolutionary Ecology and Wildlife Diseases from the Leibniz Institute for Zoo and Wildlife Research (IZW) in Berlin and the NIST in Charleston, working together Dr. Bettina Wachter, Dr. Gabor Czirjak and Dr. Ben Neely to understand the phenotypic differences between cheetahs in different life stages.
The IZW team has been studying wild cheetahs roaming Namibian farmland since 2001, focusing on developing sustainable ways to resolve cheetah-farmer conflict. An important solution lies in understanding the unique social system of cheetahs.
As previously discovered by the IZW team, cheetah males either defend small territories or roam in large home ranges covering several territories (“floaters”) [1]. These territories are not adjacent but are separated by around 23 km [2].
Floaters try to take over a territory from a resident male because they get preferential access to females. When they succeeded in doing so, their behavior and phenotype changes within a couple of months. They become aggressive and their muscle mass increases significantly [1]. This is remarkable, because in other species such physiological changes always occur before the takeover of a territory, not after.
It is not known what triggers the behavioral and physiological changes in floaters once they became territorial. However, preliminary studies of the IZW demonstrated that it is not triggered by the male sexual hormone testosterone. Therefore, we delved into the proteome of the two cheetah male types to shed light into this striking change in body composition, as well as the underlying biological differences between male and female cheetahs.
Interrogating the cheetah proteome
The team of the IZW humanely captured wild cheetahs with box traps at cheetah marking trees, fitting them with GPS collars and collecting blood samples. Blood samples were transported to the research station, centrifuged and plasma samples were frozen in liquid nitrogen for transportation to the IZW.
For this study, plasma samples of 45 cheetahs were selected, including 15 females, 15 territorial males and 15 floater males, collected between 2004 and 2021. The samples were stored frozen at the IZW for up to 18 years before reaching us at Biognosys and still remained viable for analysis, showing that this approach could be used for similar research programs.
Once we received the samples at Biognosys, we set to work analyzing them using data-independent acquisition (DIA) mass spectrometry, our specialist workflow for discovery proteomics with our TrueDiscovery™ platform.
This workflow uses Hyper Reaction Monitoring (HRM™) to quantify differentially expressed proteins across the entire proteome. The method also generates peptide-level information for all detectable proteins. With our proprietary protein libraries, we can now identify almost 100,000 peptides in biofluids such as plasma.
Using this methodology, we identified almost 2,000 proteins and 20,000 peptides in plasma – the most detailed analysis of the plasma proteome of cheetahs gathered to date.
The first biological insights
Although full analysis of these exciting new results is ongoing, we can already reveal that there are interesting biological clues hidden within the cheetah proteome.
Principal component analysis (PCA), a statistical method used to analyze the information contained in large datasets, was applied to the cheetah proteome samples and could distinguish between the two cheetah male types and the females (Figure 1).
Based on the abundance of different proteins in the samples, we found significant differences between the three cheetah groups. There was particularly noticeable separation of the territorial males from the other two groups (floaters and females), which may explain the striking phenotypic differences between territorial males and floaters.
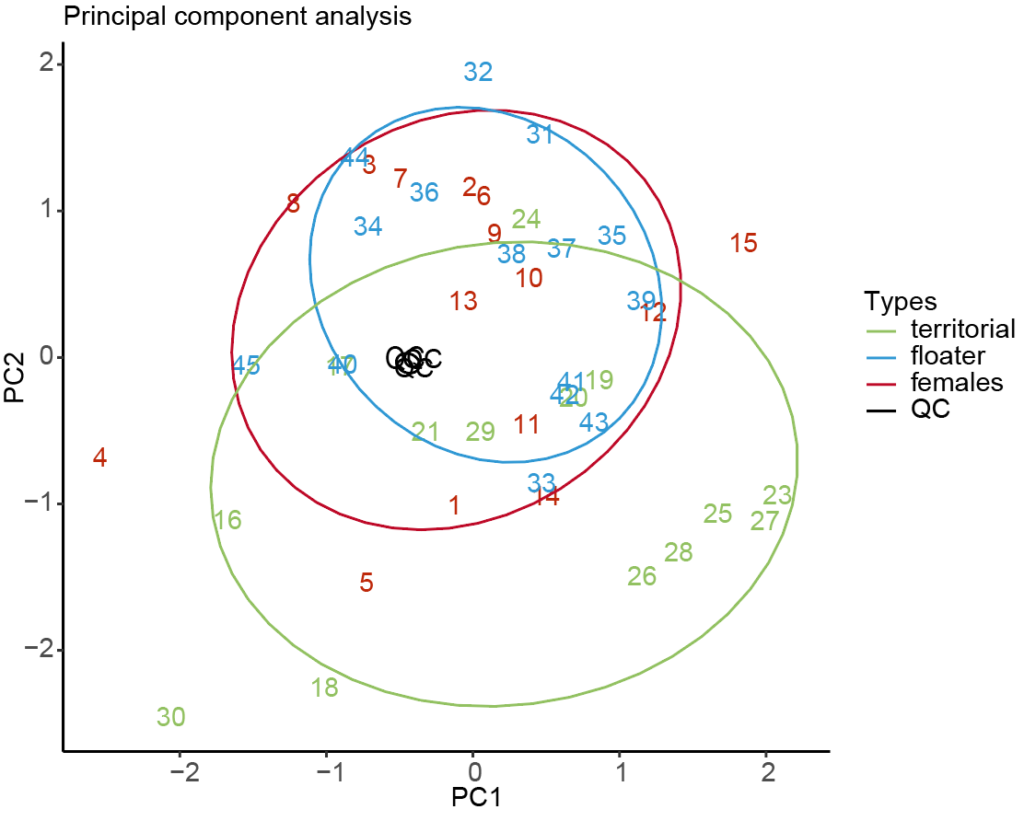
Figure 1: Principal component analysis (PCA) of the three cheetah types: territorial males, floater males and females. Workflow quality control showed low technical variance (QC).
We also saw significant differences in post-translational modification (PTMs) of the proteome samples, such as methylation and hydroxylation, which can change the properties of proteins and may explain functional differences between groups. Further research is required to understand the biological context of these modifications and how they relate to the different cheetah phenotypes.
Proteomics powering fundamental biology research
We are deeply encouraged by these findings, which are just the beginning of a much larger project. We are continuing to analyze the data and expect to make more exciting discoveries, with a paper to be published soon.
For example, we are planning to use proteomics to study the extraordinary immune system of wild cheetahs. All of the 500+ wild cheetahs trapped and released by the IZW team over the years are clinically healthy and have a very strong constitutive immune system, despite being infected with various pathogens [3,4,5]. In contrast, cheetahs living in zoos often show clinical symptoms when infected with pathogens.
Comparing the immune proteomes of wild and captive cheetahs could help us to understand what helps the wild cheetah fend off disease so capably, with potential applications in animal and human health.
We believe cutting-edge proteomics approaches hold much promise for animal research, both in laboratory and wild animals to support fundamental biology and conservation research.
More than this, discovery proteomics is a key technology for research in life sciences. There are many important discoveries waiting to be made in the deep ocean of the proteome, opening up new avenues of exploration and understanding of how life works.
At Biognosys, our next-generation platform for discovery proteomics – TrueDiscovery™️ – is the only technology that searches the complete proteome to reproducibly identify and quantify thousands of proteins at scale.
Whatever your research species and wherever in the world they live, get in touch today to discuss how our next-generation proteomics tools can unlock their biological secrets.
References
- Melzheimer J, Streif S, Wasiolka B, Fischer M, Thalwitzer S, Heinrich SK, Weigold A, Hofer H, Wachter B 2018: Queuing, take-overs, and becoming a fat cat: Long-term data reveal two distinct male spatial tactics at different life-history stages in Namibian cheetahs. Ecosphere 9(6): e02308. Doi: 10.1002/ecs2.2308.
- Melzheimer J, Heinrich SK, Wasiolka B, Mueller R, Thalwitzer S, Palmegiani I, Weigold A, Portas R, Roeder R, Krofel M, Hofer H, Wachter B 2020: Communication hubs of an asocial cat are the source of a human-carnivore conflict and key to its solution. Proceedings of the National Academy of Sciences of the United States of America 117(52), 33325-33333. www.pnas.org/cgi/doi/10.1073/pnas.2002487117.
- Thalwitzer S, Wachter B, Robert N, Wibbelt G, Müller T, Lonzer J, Meli ML, Bay G, Hofer H, Lutz H 2010. Seroprevalences to viral pathogens in free-ranging and captive cheetahs (Acinonyx jubatus) on Namibian farmland. Clinical and Vaccine Immunology 17: 232-238. Doi:10.1128/CVI.00345-09.
- Heinrich SK, Wachter B, Aschenborn OHK, Thalwitzer S, Melzheimer J, Hofer H, Czirják GÁ 2016: Feliform carnivores have a distinguished constitutive innate immune response. Biology Open 5: 550-555. Doi: 10.1242/bio.014902.
- Heinrich SK, Hofer H, Courtiol A, Melzheimer J, Dehnhard M, Czirják GÁ*, Wachter B* 2017: Cheetahs have a stronger constitutive innate immunity than leopards. Scientific Reports 7:44837. Doi: 10.1038/srep44837.* Authors contributed equally to this work.

