Proteomics Meets Health Data Analytics
Advances in data science enable proteome data at a large scale to be combined with other biodata and electronic patient health records, generating revolutionary novel insights for biomedical research and patient care.
The proteome is a rich and sensitive indicator of the internal state of a cell or an organism at any particular moment. Advances in technology and analytical capabilities over recent years have brought proteomics into the spotlight as a robust, quantitative tool for measuring health and disease.
By studying the whole proteome, we can uncover aberrant molecular pathways, characterize the action of new drug candidates and identify novel biomarkers.
Further power will come from using cutting-edge data science to combine proteomic data with multi-omic data and electronic health records for health data analytics. This will allows us to make more accurate diagnoses and treatments, opening the door to true precision medicine.
Mass spectrometry opens the door to whole proteome discovery
Mass spectrometry is a highly sensitive technique for detecting and measuring individual proteins, including their functional modifications, in a large variety of biological matrices. It is the only method that identifies proteins by their sequence.
The latest techniques can simultaneously identify and quantify thousands of proteins at high speed with high precision, measuring their differential expression and offering unbiased discovery across the whole proteome.
Over the past decade, the sensitivity of this technology has increased significantly. In 2019, researchers at Biognosys demonstrated that it was possible to quantify more than 10,000 proteins in a single experiment.
At the same time, improvements in standardization and automation, pioneered by Biognosys, have transformed mass spectrometry into a robust, reproducible methodology for analyzing multiple samples from large clinical trials in parallel, opening up the possibility for high-throughput proteomics.
From genotype to phenotype: proteomics as a complement to genomics
With the advent of next-generation sequencing (NGS), we now have access to vast volumes of genomic data from a growing number of individuals.
However, gaining insights into health and disease has remained challenging due to the enormous complexity of the genome and its inherently static nature.
Being able to identify genetic variants in the genome reveals little about the state of health at the cellular or organismal level, and of the millions of genetic variants cataloged to date, less than 0.2% have been fully characterized.
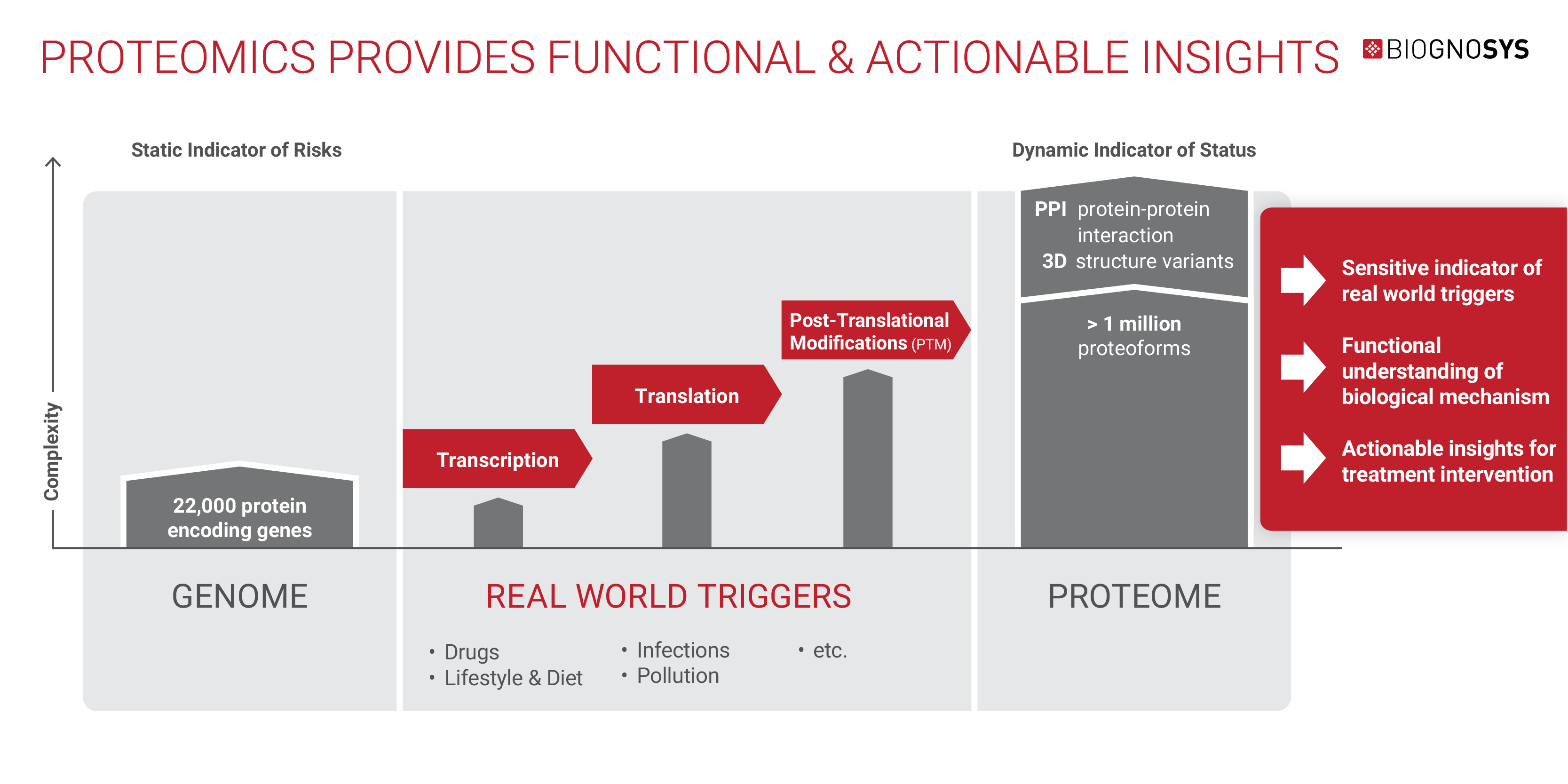
The proteome effectively responds to the internal and external environment of the body, making it a sensitive indicator of factors that influence health, such as diet, habits, clinical history, and the outside world.
While the human proteome is also highly complex, with splicing variants and post-translational modifications generating more than a million different proteoforms from around 22,000 genes, there are significant benefits to using proteomics to probe health and disease states.
The proteome provides valuable functional information, revealing the phenotypic state of cells and tissues on a molecular level. It also provides actionable information about modifiable targets and pathways potentially involved in disease, along with biomarkers that can act as a readout of health status.
Modern high-throughput mass spectrometry techniques allow all this rich data to be uncovered at scale in an unbiased way across the whole proteome.
Building precision proteomics workflows for robust comparisons
The increasing sensitivity of mass spectrometry methods to identify low abundance proteins in complex samples such as tumors or blood plasma has sparked interest in the application of proteomics techniques across many areas of biomedical research.
However, due to the highly variable nature of the proteome in response to internal and external changes, it was difficult in the past to compare multiple samples and time points.
In recent years, calibration and standardization methods for mass spectrometry have evolved significantly, from taking and processing samples through to technical workflows and data analysis.
These advances finally allow robust longitudinal data comparability across thousands of patient samples, identifying and monitoring novel biomarkers and generating new clinical insights.
Connecting proteomics to health records
Many health services worldwide are using electronic health records (EHRs) to store medical information about individuals. These records are a rich source of health data for research into the origins and treatment of disease.
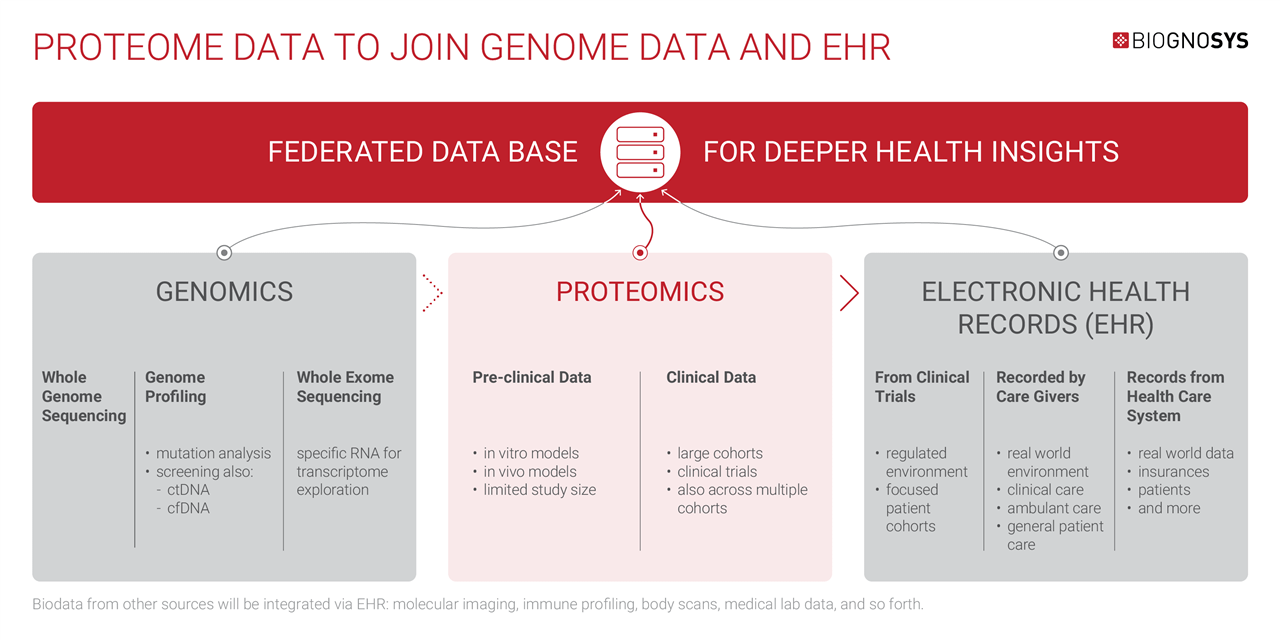
Increasingly, researchers are matching genomic data to health records to gain insights into the risks and causes of various conditions. However, given that the proteome provides a highly sensitive readout of the body’s current internal state, it is likely that aligning proteomic data to EHRs will reveal far more information about health and well-being than genomics alone.
In the future, EHRs will become the central hub for all kinds of personal biodata gathered throughout an individual’s lifetime, including proteomics, genomics, imaging, histology, lab tests, and more, creating a truly multi-modal health repository.
Biognosys is working with our partners and clients to develop new standards and tools that can integrate proteomic data with other forms of health data for complex analysis, making this vision a reality.
Seamless data analytics requires large scale approaches
Gathering sufficient data together will need seamless long-term collaboration across a network of trusted partners. They will provide suitable data and samples from large-scale and ideally longitudinal studies and additional sources.
On the proteomics side, two components must come together: the capacity to generate large-scale datasets in high-throughput proteomics analysis and highly standardized instrumentation delivering coherent, high-quality data over time. To enable this, Biognosys has invested in our large-scale facility in Switzerland, which will soon have the capacity to analyze hundreds of thousands of samples a year.
We are also committed to making proteomic data as useful as possible, building competency in health data analytics, and supporting long-term access and re-use of data for any distinct research context.
Putting proteome data at the heart of biodata
We are now at a point where proteome data is ready to become a standard part of health data analysis. Proteome data will be aligned with biodata from other sources, such as genomic profiling, imaging, histology, and so forth, to create a truly multi-modal view of life. Matching proteome data against EHRs will give us unprecedented insights into human health.
Biognosys is committed to realizing the benefits of combining proteomics with advanced health data analytics, supporting our customers and partners to integrate and analyze complex datasets. Not only do we provide you with robust, large-scale proteomics data, but we will work together with you to unlock the novel clinical insights within it.
We invite you to join us on this fascinating journey. Get in touch with us to start the conversation. And for more information about Biognosys, explore our website here.

